Content from Introduction
Last updated on 2025-05-06 | Edit this page
Overview
Questions
- What is the NextGen Framework?
- What is NextGen in a Box (NGIAB)?
- What is containerization?
- Why should I use NGIAB?
Objectives
- Identify key components of the NGIAB architecture
- Describe NGIAB’s role in the NextGen Framework
- Determine use cases for NGIAB
Introduction to NextGen
The U.S. National Water Model (NWM) provides hydrologic predictions for over 2.7 million river reaches across the United States (Cosgrove et al., 2024). The Next Generation Water Resources Modeling Framework (NextGen) is an advancement of the NWM, setting the stage for a more flexible modeling approach. NextGen promotes model interoperability and standardizes data workflows, allowing the integration of various hydrologic models tailored to specific regional processes, providing key flexibility needed for future success with continental-scale modeling. The NextGen framework continues to undergo testing, improvements, and updates through research efforts at the NOAA Cooperative Institute for Research to Operations in Hydrology (CIROH).
What is NGIAB?
Managing NextGen’s complex software ecosystem remains challenging. The NextGen framework’s implementation requires handling numerous software libraries and dependencies. To streamline this, we developed NextGen In A Box (NGIAB)—an open-source, containerized solution that encapsulates the NextGen framework and essential modeling components into a self-contained, reproducible application. By eliminating manual configuration burdens, NGIAB enables researchers to focus on scientific inquiry rather than software setup and maintenance. Beyond simplifying deployment of the NextGen Framework, NGIAB fosters collaboration among researchers, academic institutions, and government agencies by providing a scalable, community-driven modeling environment. In essence, NGIAB provides a unified solution that powers NextGen models, including future versions of the NWM starting with version 4.
Terminology
- NextGen: overarching framework
- NGIAB: containerized packaging of NextGen
-
ngen: the engine used within NextGen - NWM: specific operational implementation used by the National Weather Service
Containerization
- Containerization addresses compatibility issues and hardware variation challenges by encapsulating applications, their dependencies, and runtime environments into a single, portable unit.
- Think of containerization like putting a model and all its tools into a sealed toolbox – you can carry and run it anywhere, and everything needed is inside..
- This ensures consistent execution across diverse computing environments, regardless of differences in hardware or software configurations.
- NGIAB leverages Docker (Boettiger, 2015) and Singularity (Hunt et al., 2005) to streamline deployment.
Architectural Components
NGIAB is designed as a multi-layered containerized tool that encapsulates the NextGen framework and many components relevant to the NWM within a reproducible environment.
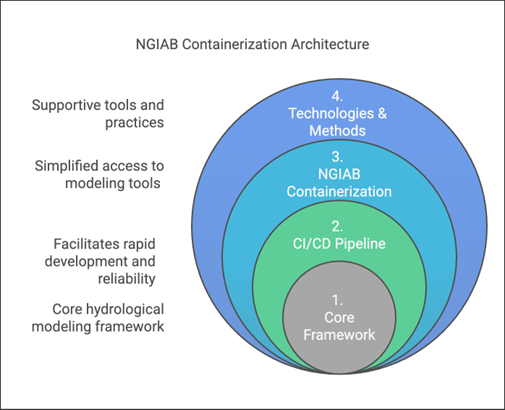
Figure 1 illustrates the layered architecture of NGIAB.
- Layer 1: At its core (Layer 1) lies a suite of integrated hydrological modeling components and hydrofabric (a geopatial dataset representing hydrologic features like rivers, basins, and connections), designed to work together within the NextGen framework. Hydrologic models in NGIAB are Basic Model Interface (BMI) compliant, meaning that they follow a standard structure and can be swapped in and out for one another.
- Layer 2: Layer 1 is wrapped by the Continuous Integration/Continuous Deployment (CI/CD) Pipeline layer (Layer 2). CI/CD are tools and practices that automate code testing and updates. NGIAB leverages GitHub Actions to ensure automated testing, integration, and deployment capabilities for reproducible workflows.
- Layer 3: The NGIAB Containerization layer (Layer 3) provides the containerized environment and essential configuration tools.
- Layer 4: The outermost layer (Layer 4), Technologies & Methods, provides broader infrastructure, best practices, and support for deployment across different computing environments (local, cloud, HPC), and facilitates community engagement and contribution.
The architecture emphasizes four key aspects:
- core hydrological modeling framework capabilities,
- simplified access to modeling tools,
- facilitation of rapid development and reliability,
- and integration of supportive tools and practices.
Extensions of NGIAB
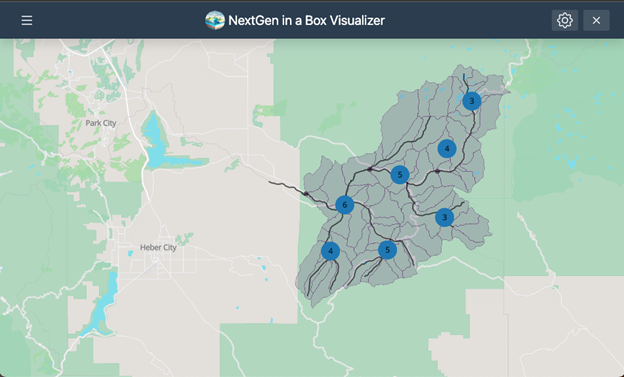
Several extensions of NGIAB are already integrated with NextGen-related tools like Data Preprocess, Tools for Exploratory Evaluation in Hydrologic Research (TEEHR), and Data Visualizer (Figure 2). These extensions will be discussed in later episodes.
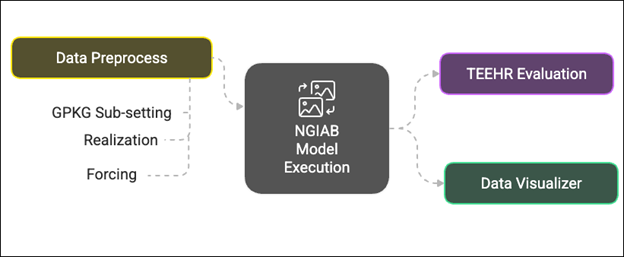
Example Applications
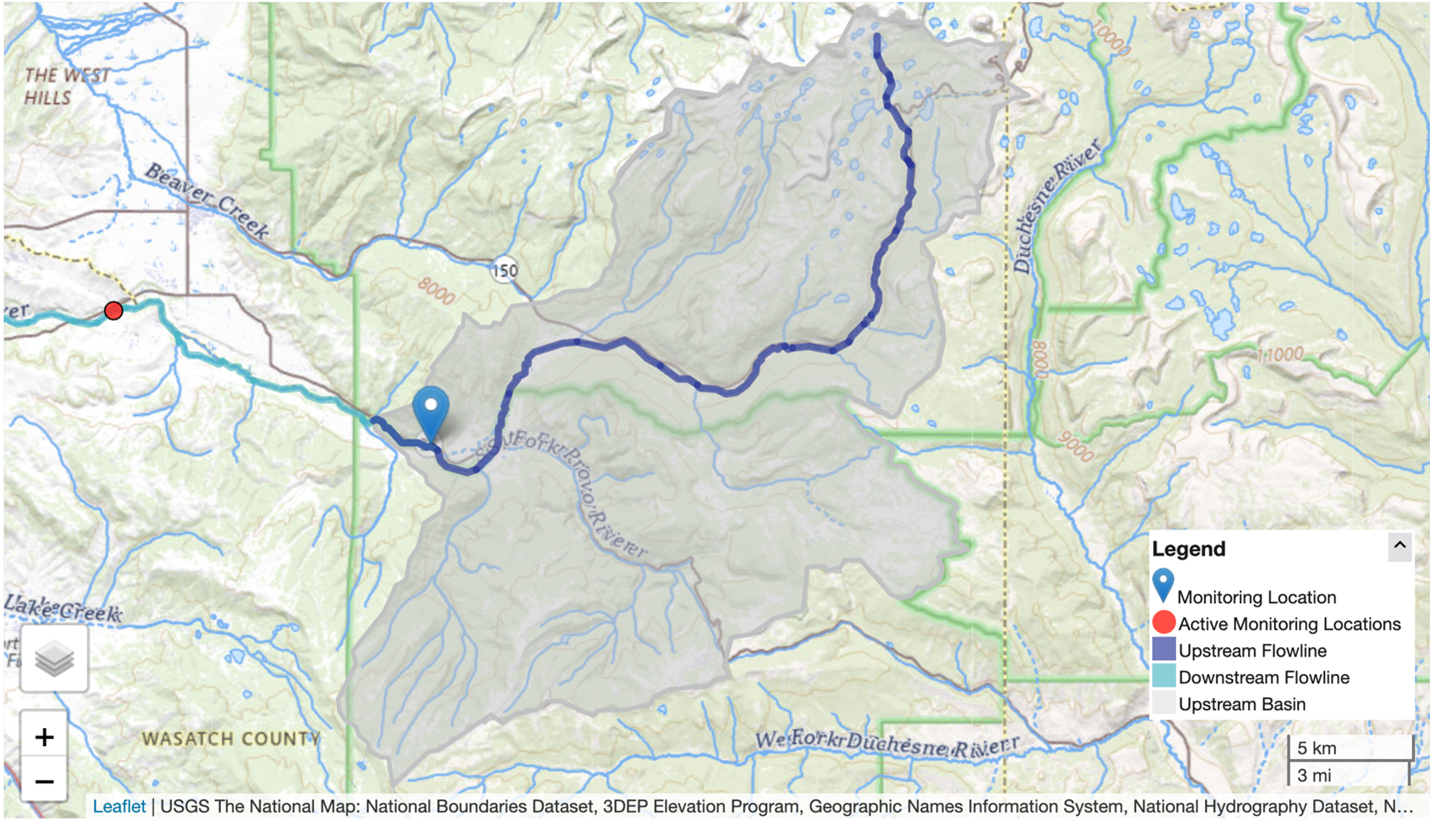
Steps common to all hydrologic modeling frameworks include data collection and preparation, framework setup and model execution, evaluation, results visualization, and calibration. Researchers can use NGIAB to execute model runs for their basins of interest. Note that calibration is not yet an integrated capability within NGIAB. Figures 3 and 4 show examples of how NGIAB and its extensions have been used to simulate streamflow for five years in the Provo River basin.
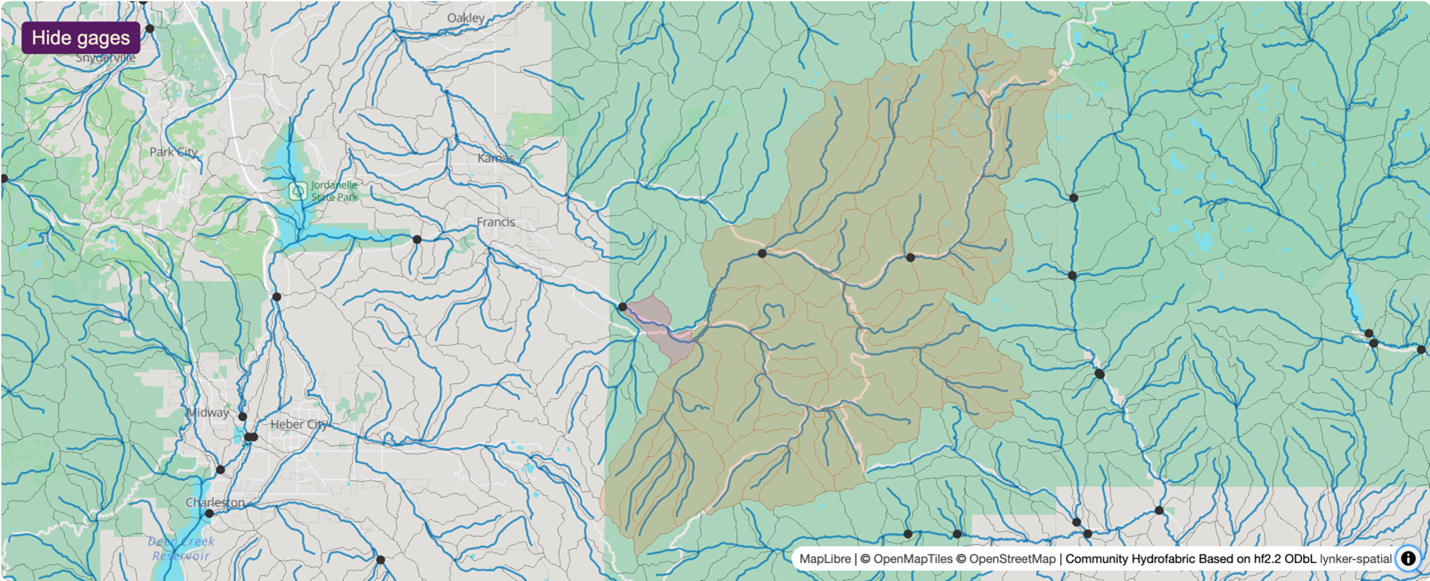
Why should I use NGIAB?
NGIAB makes community contribution possible in
research settings by simplifying setup and providing demos, allowing
hydrologists and researchers to configure and modify localized water
models. Built on open-source code and the ngen/BMI
foundation, NGIAB allows integration of a hydrology process model into a
larger hydrologic simulation framework, allowing a researcher to focus
on their area of specific modeling expertise. Its lightweight container
size also empowers hydrologists to execute large-scale runs efficiently
and reduce computational bottlenecks. By strengthening collaboration
across research teams, NGIAB will help drive the evolution of
community-scale water modeling and accelerate the transition from
academic innovation to real-world operational use.
Your Turn
Here are some self-assessment questions for discussion or consideration:
- Do I understand how NGIAB fits into the NextGen Framework?
- What are the key design features and extensions of NGIAB?
- How can I use NGIAB to answer my research questions?
- How can I use NGIAB to contribute my expertise to the NextGen Framework?
- The Next Generation Water Resources Modeling Framework (NextGen) advances the National Water Model with flexible, modular, and regionally adaptive hydrologic modeling at national scale.
- NextGen In A Box (NGIAB) packages the complex NextGen system into an open-source, containerized application for easier access and usability.
- NGIAB uses Docker and Singularity for portability across local machines, cloud platforms, and HPC systems.
- NGIAB’s multi-layered architecture integrates hydrologic modeling tools, CI/CD pipelines, and supportive technologies and is complemented by a suite of extensions that allow for end-to-end workflows from data acquisition to visualization and evaluation.
- NGIAB fosters an open ecosystem where researchers, developers, and practitioners actively contribute new models, extensions, and workflows.
Content from Installation and Setup
Last updated on 2025-05-23 | Edit this page
This episode can be a standalone tutorial for those who want a quick introduction to NGIAB. This tutorial follows the case study from our CloudInfra repository. Users who wish to learn more about NGIAB can explore our other episodes in this module.
Overview
Questions
- How do I install and set up NGIAB?
- What are the prerequisites for running NGIAB?
- How do I verify my installation?
Objectives
- Install and verify Docker
- Set up NGIAB project directories
- Run a sample NGIAB run
Introduction
This lesson guides you through installing and setting up NGIAB, a containerized solution designed to simplify running the NextGen modeling framework locally. NGIAB leverages Docker containers to ensure consistent and reproducible runs.
Are you using an HPC?
Instead of following these instructions, follow the guidance in the HPC sections in Advanced Topics.
System Requirements
Before installing NGIAB, ensure you have:
-
Operating System: Windows (with WSL), macOS, or
Linux
-
Software: Docker, Git
- Recommended Minimum RAM: 8 GB
Connecting to a remote machine through SSH?
To use the Data Visualizer through an Secure Shell (SSH) connection, you will have to set up port forwarding when connecting to the remote machine. Port forwarding will allow you to access a remotely hosted browser session on your local machine. See the instructions under “Using NGIAB through an SSH connection” in the Advanced Topics episode in this training module.
Docker Installation
Note: Users who already have Docker installed will still need to install a separate WSL distro and set it as their default, if they have not already.
-
Install Windows Subsystem for Linux (WSL):
Install Docker Desktop from Docker’s official website.
Launch Docker Desktop and open WSL terminal as administrator.
-
Verify Docker installation:
This should generate a message that shows that your installation is working.
-
Install Astral UV:
CAUTION: WSL distributions
NGIAB commands cannot be run through the docker-desktop
distribution. If you installed Docker before WSL, you will likely need
to install a new WSL distribution and set it as your default.
For example, Ubuntu can be installed and set as the default distribution with the following commands:
Install Docker Desktop from Docker’s official Mac installer.
Launch Docker Desktop.
-
Verify Docker installation:
This should generate a message that shows that your installation is working.
-
Install Astral UV.
Install Docker by following the official Docker guide.
-
Start Docker service and verify:
This should generate a message that shows that your installation is working.
-
Install Astral UV.
NGIAB Setup
These steps will lead you through the process of running NGIAB with a set of pre-configured input data and realization files. A realization file is a scenario using a specific model on a specific region.
Step 2: Download Sample Data
Choose one of the following datasets. File sizes and model configurations are provided so that you can download a dataset suitable for your interests, available disk space, and network speed.
Option 1: AWI-009
Models: SLOTH (dummy model), NoahOWP (land surface model), CFE (conceptual rainfall-runoff model, functionally equivalent to NWM)
Compressed file size: 249 MB | Extracted file size: 1.77 GB
Option 2: AWI-007
Models: SLOTH, NoahOWP, CFE
Compressed file size: 1.87 MB | Extracted file size: 5.53 MB
Step 3: Clone and Run NGIAB
CAUTION: For Windows users: pulling with LFs
Before cloning the repository, please ensure that Git is configured to pull with LF line breaks instead of CRLFs. If CRLFs are used, then the carriage return characters will prevent the shell scripts from running properly.
There are a couple options to configure this.
Visual Studio Code can be used to manually toggle between line breaks.
Git can be configured from the command line.
- Download, extract, and run this interactive
.batscript
✅ Ready to Go!
If you’ve completed the steps above and verified your dataset and
working directory, you are ready to run the interactive guide script
guide.sh. It will prompt you to select input data,
processing modes, and will initiate your run.
This will walk you through the NGIAB setup and launch your first run.
guide.sh Tips
- A series of prompts will appear that ask you if you want to use the existing Docker image or update to the latest image. Updating to the latest image will take longer, so for the purposes of this tutorial, using the existing Docker image is fine.
- When prompted to run NextGen in serial or parallel mode, either is fine.
- The option to open a Bash shell (interactive shell) will allow you to explore the data directory without quitting NGIAB.
- Redirecting command output to
/dev/nullsignificantly reduces the amount of output. Either is fine, but if you are curious about what is happening inside the model, we suggest that you do not redirect the output.
Troubleshooting
Ensure Docker is running before executing
guide.sh.For permission errors on Linux, run Docker commands with
sudoor add your user to the Docker group:
Tip: Always execute a quick run with provided sample datasets to verify the successful setup of NGIAB.
Additional Resources
Are you interested in customizing your run with your own catchments (watersheds) and run configurations? Do you want to explore more functionalities of NGIAB? Check out the following episodes:
- NGIAB simplifies NextGen framework deployment through Docker.
- Use
guide.shfor interactive configuration and run execution.
- Always confirm successful setup by executing provided sample runs.
Content from Data Preparation
Last updated on 2025-05-09 | Edit this page
Overview
Questions
- How should I prepare my run directory?
- What is the Data Preprocess tool?
Objectives
- Identify the required data structure of a NextGen run in NGIAB
- Explain how the Data Preprocess tool interacts with NGIAB
- Prepare data for an NextGen run in NGIAB
Data Preprocess Tool
The Data Preprocess tool streamlines data preparation for NextGen runs in NGIAB. This tool provides a graphical user interface (GUI) and a command line interface (CLI) to prepare input data and execute model runs. A graphical user interface facilitates catchment and date range selection options via an interactive map, simplifying the subsetting of hydrofabrics, generation of forcings, and creation of default NextGen realizations. While this module reduces procedural complexity, it incorporates pre-defined assumptions that may limit user flexibility in specific applications (Cunningham, 2025).
Installing and Using the Data Preprocess Tool
The Data Preprocess tool (like all of our software) is constantly being updated and refined. As of the time of writing (see last updated date above), there are two ways to run the tool. Instructions for installation, environment management, and the GUI/CLI are found on the Data Preprocess GitHub page. We will cover some examples of the CLI usage, but full documentation is on the GitHub page.
Example 1
This command allows you to run the Data Preprocess CLI tool without installing it. It produces forcings and a NextGen realization file for the catchments upstream of gage-10154200 for the time period 2017-09-01 to 2018-09-01. Forcing data is sourced from the zarr files in the Analysis of Record for Calibration (AORC) dataset, which allows for a faster processing time.
Astral UV is required to run the Data Preprocess tool without installing it.
BASH
uvx --from ngiab_data_preprocess cli -i gage-10154200 -sfr --start 2017-09-01 --end 2018-09-01 --source aorcuvx --from ngiab_data_preprocess cli indicates that the
Data Preprocess tool will run without the user installing it. The
--source flag determines where the Data Preprocess tool
will pull forcing data from.
Example 2
This command produces forcings and a NextGen realization file for the catchments upstream of gage-10155000 for the time period 2022-08-13 to 2022-08-23 after installing the Data Preprocess tool. The forcing data source defaults to the NetCDF files in the NWM 3.0 retrospective. Using this command requires you to have Astral UV (a package installer and environment manager) installed. Instructions for installing Astral UV are found in the Astral UV documentation.
To install the Data Preprocess tool, follow the latest instructions on the Data Preprocess GitHub page.
uv run cli indicates that the Data Preprocess CLI within
your activated Astral UV environment will run. The -i flag
indicates the ID of the feature that is used to subset
the hydrofabric. The -sfr flags indicate that the Data
Preprocess tool will subset the hydrofabric to the
desired catchments, produce forcings over the desired
area and time period, and produce a NextGen realization
file. The --start and --end flags indicate the
start and end dates of the desired time period.
Example 3
This command allows you to run the Data Preprocess CLI tool from a
regular pip install ngiab_data_preprocess. However,
using Astral UV is highly recommended for its speed. This
command produces forcings for the catchments upstream of cat-7080 for
the time period 2022-01-01 to 2022-02-28.
python -m ngiab_data_cli indicates that the Data
Preprocess CLI tool will execute.

NextGen Run Directory Structure (ngen-run/)
Running NextGen requires building a standard run directory complete with only the necessary files. This is done automatically with the Data Preprocess tool. Below is an explanation of the standard run directory.
A NextGen run directory ngen-run contains the following
subfolders:
-
config: model configuration files and hydrofabric configuration files. (required) -
forcings: catchment-level forcing timeseries files. Forcing files contain variables like wind speed, temperature, precipitation, and solar radiation. (required) -
lakeout: for t-route (optional) -
metadataprogrammatically generated folder used within ngen. Do not edit this folder. (automatically generated) -
outputs: This is where ngen will place the output files. (required) -
restart: For restart files (optional)
ngen-run/
│
├── config/
│
├── forcings/
│
├── lakeout/
|
├── metadata/
│
├── outputs/
│
├── restart/Configuration directory ngen-run/config/
This folder contains the NextGen realization file, which serves as the primary model configuration for the ngen framework. This file specifies which models to run (such as NoahOWP/CFE, LSTM, etc), run parameters like date and time, and hydrofabric specifications (like location, gage, catchment).
Based on the models defined in the realization file, BMI configuration
files may be required. For those models that require per-catchment
configuration files, a folder will hold these files for each model in
ngen-run/config/cat-config. See the directory structure
convention below.
ngen-run/
|
├── config/
| │
| ├── nextgen_09.gpkg
| |
| ├── realization.json
| |
| ├── ngen.yaml
| |
| ├── cat-config/
| │ |
| | ├──PET/
| │ |
| | ├──CFE/
| │ |
| | ├──NOAH-OWP-M/NextGen requires a single geopackage file. This file is the hydrofabric (Johnson, 2022) (spatial data). An example geopackage can be found on Lynker-Spatial’s website. Tools to subset a geopackage into a smaller domain can be found at Lynker’s hfsubset.
Your Turn
Using the Data Preprocess tool, you should be able to create a run directory for your desired catchment that can be used with NGIAB. Try out both the GUI and the CLI, and experiment with different arguments and selection tools!
-
ngen-run/is the standard NextGen run directory, containing the realization files that define models, parameters, and run settings; forcing data; outputs; as well as the spatial hydrofabric. - The Data Preprocess tool simplifies preparing data for NextGen by offering a GUI and CLI for selecting catchments and date ranges, subsetting hydrofabric data, generating forcing files, and creating realization files.
Content from Model Execution
Last updated on 2025-05-01 | Edit this page
Overview
Questions
- How do I execute a NextGen run?
Objectives
- Recognize methods to execute NextGen models
- Execute a NextGen run using NGIAB
Model Execution using guide.sh
guide.sh is used to execute pre-configured NextGen runs
in NGIAB. These settings can be configured by users ahead of time using
the Data Preprocess
tool. Execute the following commands:
BASH
cd NextGen
git clone https://github.com/CIROH-UA/NGIAB-CloudInfra.git
cd NGIAB-CloudInfra
./guide.shguide.sh will prompt you to enter input data pathways
and allow you to select a computational mode (serial or parallel
processing). After the run is complete, guide.sh will give
you the option to evaluate model predictions and visualize results
(discussed in the next two episodes).
Model Execution using Data Preprocess tool
A secondary method for executing a NextGen run in NGIAB is by using
the Data Preprocess tool’s CLI. The -a argument in the
command will automatically run NGIAB after preprocessing selected data.
As this module is being updated constantly, check back on its GitHub page
for the latest updates on its functionality.
Your Turn
Use guide.sh to execute a NextGen run in NGIAB using
your preprocessed data.
Extra Credit: Use the Data Preprocess tool to automatically execute a NextGen run in NGIAB.
- To execute a NextGen run in NGIAB with full functionality, use
guide.shin the NGIAB container. - A NextGen run in NGIAB can also be automatically executed post-preprocessing using the Data Preprocess tool.
Content from Evaluation
Last updated on 2025-04-25 | Edit this page
Overview
Questions
- How do I use Tools for Exploratory Evaluation in Hydrologic Research (TEEHR) to evaluate my NextGen models in NGIAB?
Objectives
- Explain how TEEHR interacts with NGIAB
- Evaluate a NextGen run using TEEHR in NGIAB
Using Tools for Exploratory Evaluation in Hydrologic Research (TEEHR) with NGIAB
A TEEHR evaluation run is executed by default upon execution of
guide.sh. A separate runTeehr.sh script is
also available in the NGIAB-CloudInfra repository.
What is TEEHR?
TEEHR is a Python-based package enabling analysis of hydrologic model performance (RTI International, 2025). TEEHR also provides tools to fetch the U.S. Geological Survey (USGS) streamflow data and NWM gridded and point-based retrospective and near real-time forecast data. This functionality supports comprehensive model evaluation and uncertainty analysis. NGIAB leverages TEEHR as a tool for hydrological model evaluation (CIROH, 2024). Researchers can explore a comprehensive range of metrics, including error statistics, skill scores, hydrologic signatures, and uncertainty quantification.
Gridded Data vs. Point Data
Gridded data consists of values at regularly spaced 2-dimensional cells that form a grid covering a region on Earth. Point data consists of values on specific 0-dimensional points. The NWM real-time forecast produces point-type stream routing and reservoir variables.
TEEHR folder contents
TEEHR consolidates data from the USGS and NWM, allowing side‐by‐side visual comparisons of observed and simulated over the model run intervals. Figure 1 shows the default comparison of the modeled outlet hydrograph and the corresponding time series from the NWM 3.0. While the default configuration produces this view, the TEEHR user documentation provides additional examples of capabilities for customized plotting functions.
![A hydrograph spanning years 2017-2022. The x-axis is labeled "Datetime", and the y-axis is labeled "streamflow_hourly inst [m^3/s]". The blue line represents the NextGen run (labeled "ngen"), and the orange line represents the NWM 3.0 time series (labeled "nwm30_retrospective"). A legend is in the upper-right corner explaining the colors of these lines.](fig/fig5-1.png)
guide.sh script included with NGIAB and is named
timeseries_plot_streamflow_hourly_inst.html in the
teehr folder.In addition to hydrograph visualization, TEEHR also calculates key
performance metrics such as Kling-Gupta Efficiency (KGE), Nash-Sutcliffe
Efficiency (NSE), and Relative Bias (RB) to quantify the accuracy of
model predictions against observed data (Gupta et al.,
2009; Nash
and Sutcliffe, 1970). Users do not need to understand all these
metrics immediately, and can refer to the references above to learn
more. These results are then stored in a standardized output directory.
For instance, the metrics.csv file in the TEEHR folder
structure contains aggregated statistics for each configuration.
Through this systematic approach, NGIAB and TEEHR together allow hydrologists and stakeholders to inspect, compare, and refine model predictions in an open-source environment.
Your Turn
Go ahead and execute the TEEHR run using guide.sh and
explore your TEEHR folder!
guide.sh Tips
- The default TEEHR image is fine to use.
- Tools for Exploratory Evaluation in Hydrologic Research (TEEHR) is a Python-based backage for hydrologic model evaluation.
- NGIAB uses TEEHR to assess model performance, comparing predictions against USGS streamflow and NWM data and calculating performance metrics.
- TEEHR runs automatically with the main
guide.shNGIAB script.
Content from Visualization
Last updated on 2025-05-27 | Edit this page
Overview
Questions
- How do I visualize my NextGen runs?
Objectives
- Explain how the Data Visualizer complements NGIAB.
- Use the Data Visualizer to visualize results of a NextGen run in NGIAB.
Data Visualizer
The Data Visualizer component developed using the Tethys Platform (Swain et al., 2015) complements NGIAB by providing an environment for geospatial and time series visualization of catchments and nexus points (locations where objects in the hydrofabric like streams or water bodies connect). Through a web-based architecture, researchers can explore hydrological data in a spatiotemporal context (CIROH, 2025). In addition to standard map-based displays, this component also supports the visualization of the TEEHR output, including tabular metrics and interactive time series plots.
Using the Data Visualizer with NGIAB
ViewOnTethys Script
Like TEEHR, the Data Visualizer can be activated upon execution of
the main NGIAB guide script, guide.sh. A separate
viewOnTethys.sh script is also available in the
NGIAB-CloudInfra repository.
Once a run is complete, users can launch the Data Visualizer through their web browser when prompted by the guide script. Although TEEHR’s outputs can be displayed within the Data Visualizer, this tool is primarily designed to provide a broad overview of model results. Users seeking TEEHR’s more advanced analysis features can still access them outside the Data Visualizer.
One of the advantages of the viewOnTethys.sh script is
that it allows the user to keep multiple outputs for the same
hydrofabric. It prompts the user if they want to use the same output
directory by renaming it and adding it to the collection of outputs or
if they want to overwrite it.
BASH
⚠ ~/ngiab_visualizer is not empty.
→ Keep (K) or Fresh start (F)? [K/F]: k
ℹ Reclaiming ownership of ~/ngiab_visualizer (sudo may prompt)…
⚠ Directory exists: ~/ngiab_visualizer/gage-10154200
→ Overwrite (O) or Duplicate (D)? [O/D]: o
✓ Overwritten ➜ ~/ngiab_visualizer/gage-10154200
Checking for ~/ngiab_visualizer/ngiab_visualizer.json...You should be able to see multiple outputs through the UI:
Visualizer Directory Organization
The Visualizer organizes data using a directory named
ngiab_visualizer. This directory stores all the outputs
generated by the user and includes a file called
ngiab_visualizer.json. This file contains metadata that
allows the Visualizer to locate and manage the outputs from different
runs executed via the ./guide.sh script. The metadata,
specific to the Visualizer, is structured as a simple JSON file. It
lists the outputs with details such as a label, the data path, and a
unique identifier. The ./ViewOnTethys.sh script handles the
creation and management of this metadata.
JSON
{
"model_runs": [
{
"label": "gage-10154200",
"path": "/var/lib/tethys_persist/ngiab_preprocess_output/gage-10154200",
"date": "2025-05-23:13:51:51",
"id": "61026834-4235-4d39-8a8e-f076a8854148",
"subset": "",
"tags": []
},
{
"label": "gage-20454200",
"path": "/var/lib/tethys_persist/ngiab_visualizer/gage-20454200",
"date": "2025-05-23:17:00:34",
"id": "68f6cf78-188c-4e86-b797-6c40ea36e0e6",
"subset": "",
"tags": []
},
{
"label": "gage-35054600",
"path": "/var/lib/tethys_persist/ngiab_visualizer/gage-35054600",
"date": "2025-05-23:17:01:10",
"id": "6d0cb736-2dac-4ea0-a3a3-ad26cd45ef36",
"subset": "",
"tags": []
}
]
}The path /var/lib/tethys_persist/ belongs to the
$HOME env variable of the container running the visualizer.
When the user runs the ./ViewOnTethys.sh, it mounts the
directory from the host at ~/ngiab_visualizer to
/var/lib/tethys_persist/ngiab_visualizer.
However, if the user wants more control, the user can copy their data
directory to ~/ngiab_visualizer on the host(not the
container) while the container is stop,
finally the user can open the
~/ngiab_visualizer/ngiab_visualizer.json on the host(not
the container), and add the specific run to the visualizer:
JSON
....
{
"label": "gage-35054600",
"path": "/var/lib/tethys_persist/ngiab_visualizer<MY_SPECIFIC_OUTPUT_DIRECTORY_NAME>",
"date": "2025-05-23:17:01:10",
"id": "6d0cb736-2dac-4ea0-a3a3-ad26cd45ef36",
"subset": "",
"tags": []
}The user can then run ./ViewOnTethys.sh script to spin
again the container or if the user wants more control and just define
the env variables and running the container
BASH
export CONFIG_FILE="$HOME/.host_data_path.conf" \
TETHYS_CONTAINER_NAME="tethys-ngen-portal" \
TETHYS_REPO="awiciroh/tethys-ngiab" \
TETHYS_TAG="latest" \
NGINX_PORT=80 \
MODELS_RUNS_DIRECTORY="$HOME/ngiab_visualizer" \
DATASTREAM_DIRECTORY="$HOME/.datastream_ngiab" \
VISUALIZER_CONF="$MODELS_RUNS_DIRECTORY/ngiab_visualizer.json" \
TETHYS_PERSIST_PATH="/var/lib/tethys_persist" \
SKIP_DB_SETUP=false \
CSRF_TRUSTED_ORIGINS="[\"http://localhost:${NGINX_PORT}\",\"http://127.0.0.1:${NGINX_PORT}\"]"BASH
docker run --rm -d \
-v "$MODELS_RUNS_DIRECTORY:$TETHYS_PERSIST_PATH/ngiab_visualizer" \
-v "$DATASTREAM_DIRECTORY:$TETHYS_PERSIST_PATH/.datastream_ngiab" \
-p "$NGINX_PORT:$NGINX_PORT" \
--name "$TETHYS_CONTAINER_NAME" \
-e MEDIA_ROOT="$TETHYS_PERSIST_PATH/media" \
-e MEDIA_URL="/media/" \
-e SKIP_DB_SETUP="$SKIP_DB_SETUP" \
-e DATASTREAM_CONF="$TETHYS_PERSIST_PATH/.datastream_ngiab" \
-e VISUALIZER_CONF="$TETHYS_PERSIST_PATH/ngiab_visualizer/ngiab_visualizer.json" \
-e NGINX_PORT="$NGINX_PORT" \
-e CSRF_TRUSTED_ORIGINS="$CSRF_TRUSTED_ORIGINS" \
"${TETHYS_REPO}:${TETHYS_TAG}"You should see something like this using docker ps
BASH
docker ps
CONTAINER ID IMAGE COMMAND CREATED STATUS PORTS NAMES
b1818a03de9b awiciroh/tethys-ngiab:latest "/usr/local/bin/_ent…" 25 seconds ago Up 24 seconds (health: starting) 0.0.0.0:80->80/tcp, [::]:80->80/tcp tethys-ngen-portalOnce it’s healthy, you can access the visualizer at:
http://localhost:${NGINX_PORT}
The ViewOnTethys.sh script automates updating
ngiab_visualizer.json and copying your model output into
~/ngiab_visualizer.
Key points
-
ViewOnTethys.shautomates adding model outputs tongiab_visualizer.jsonand syncing data to~/ngiab_visualizer. - To customize your setup, set environment variables and run the
awiciroh/tethys-ngiabDocker image manually.
NGIAB Visualizer UI
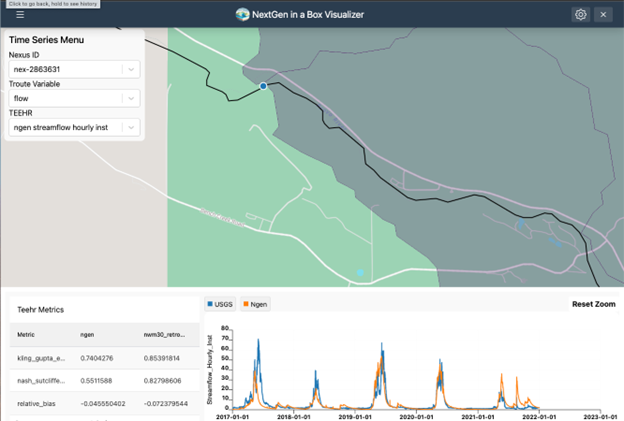
The following figures demonstrate several ways the Data Visualizer can be used to visualize model outputs, including geospatial visualization for nexus points, catchment-based visualization, and TEEHR time series representation (hydrographs).
Nexus points can be visualized when the user selects the output that wants to visualize. Time series can be retrieved by clicking on any of the Nexus points, or by changing the select dropdown assigned to the Nexus.
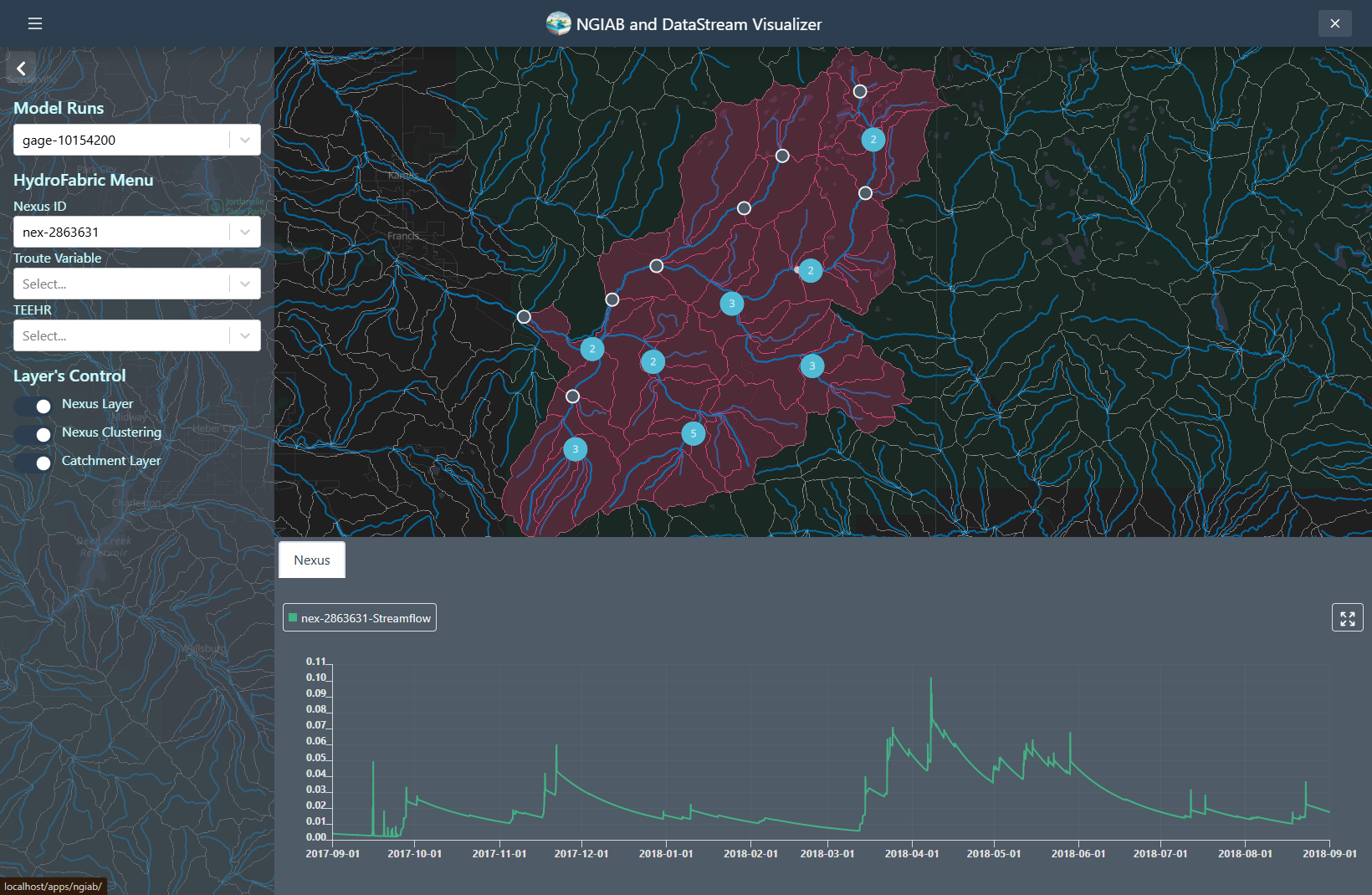
Troute variables time series can also be displayed using the Troute select dropdown.
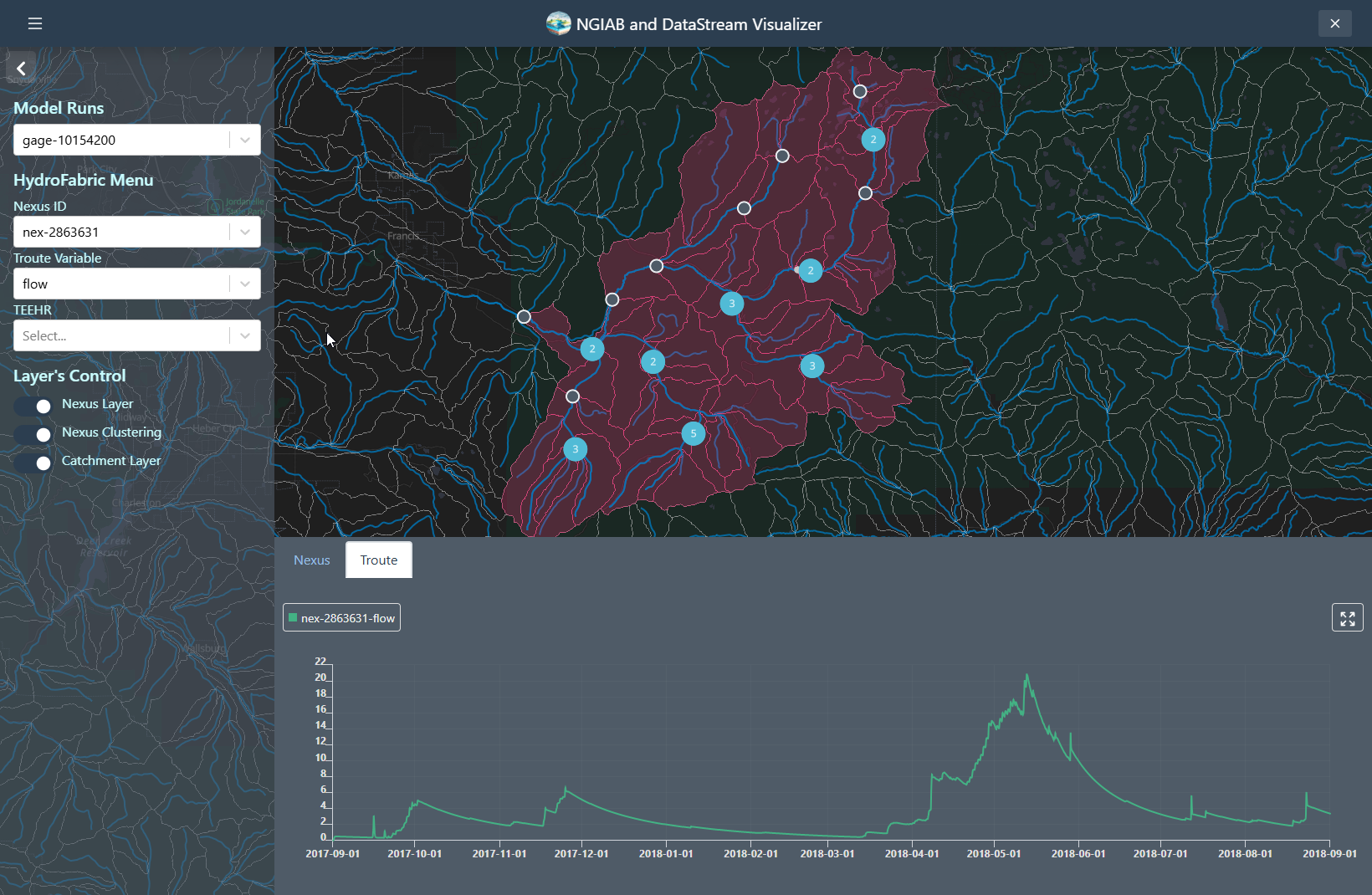
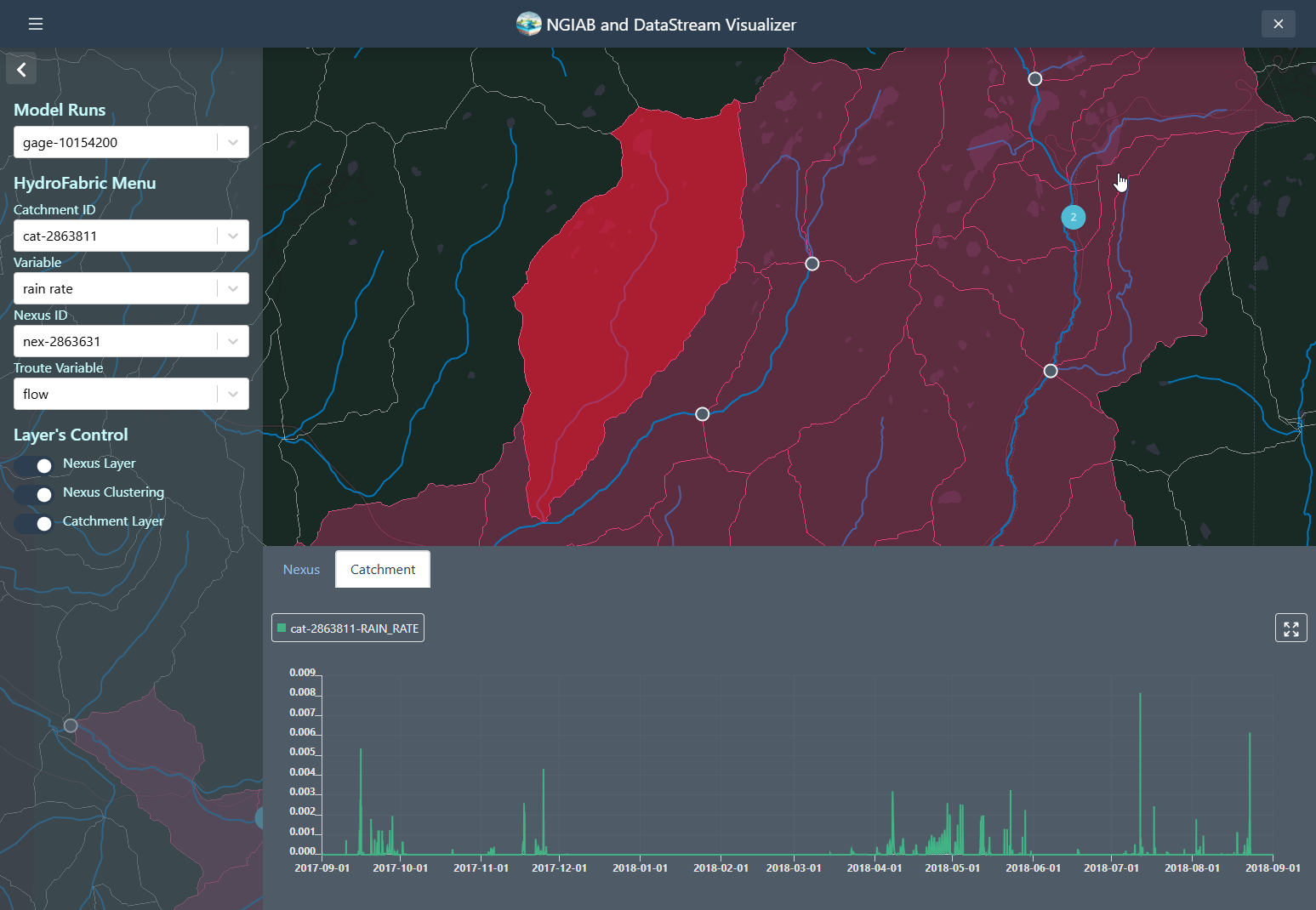
Catchments time series can be retrieved by clicking on any of the Catchments polygons, or by changing the select dropdown assigned to the Catchments.
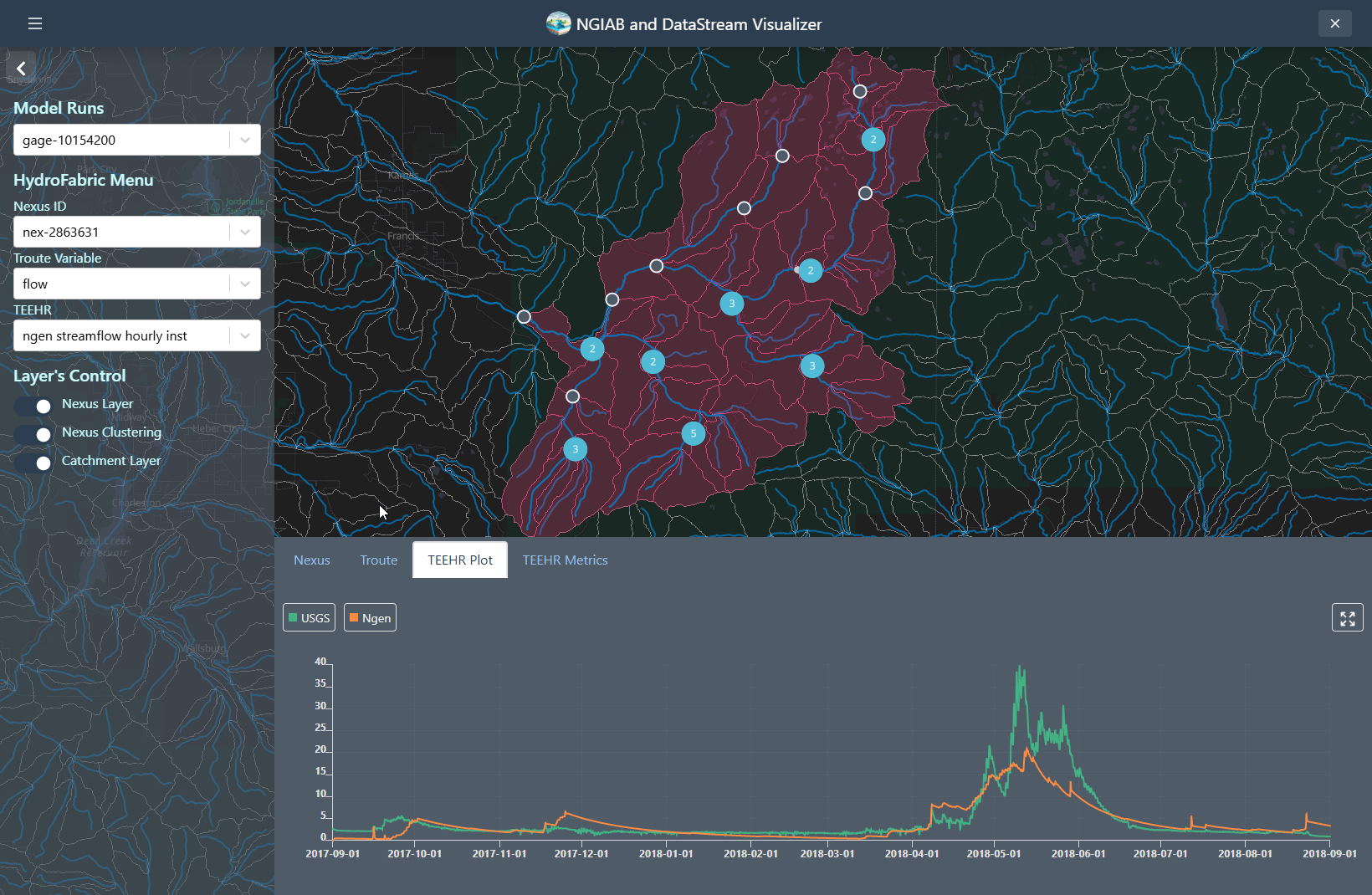
TEEHR evaluation can be visualized when the user hits a point that contains TEEHR evaluation output, the user can also look at a Nexus point on the dropdown assigned and enter the id of the Nexus points that contains TEEHR evaluation output.
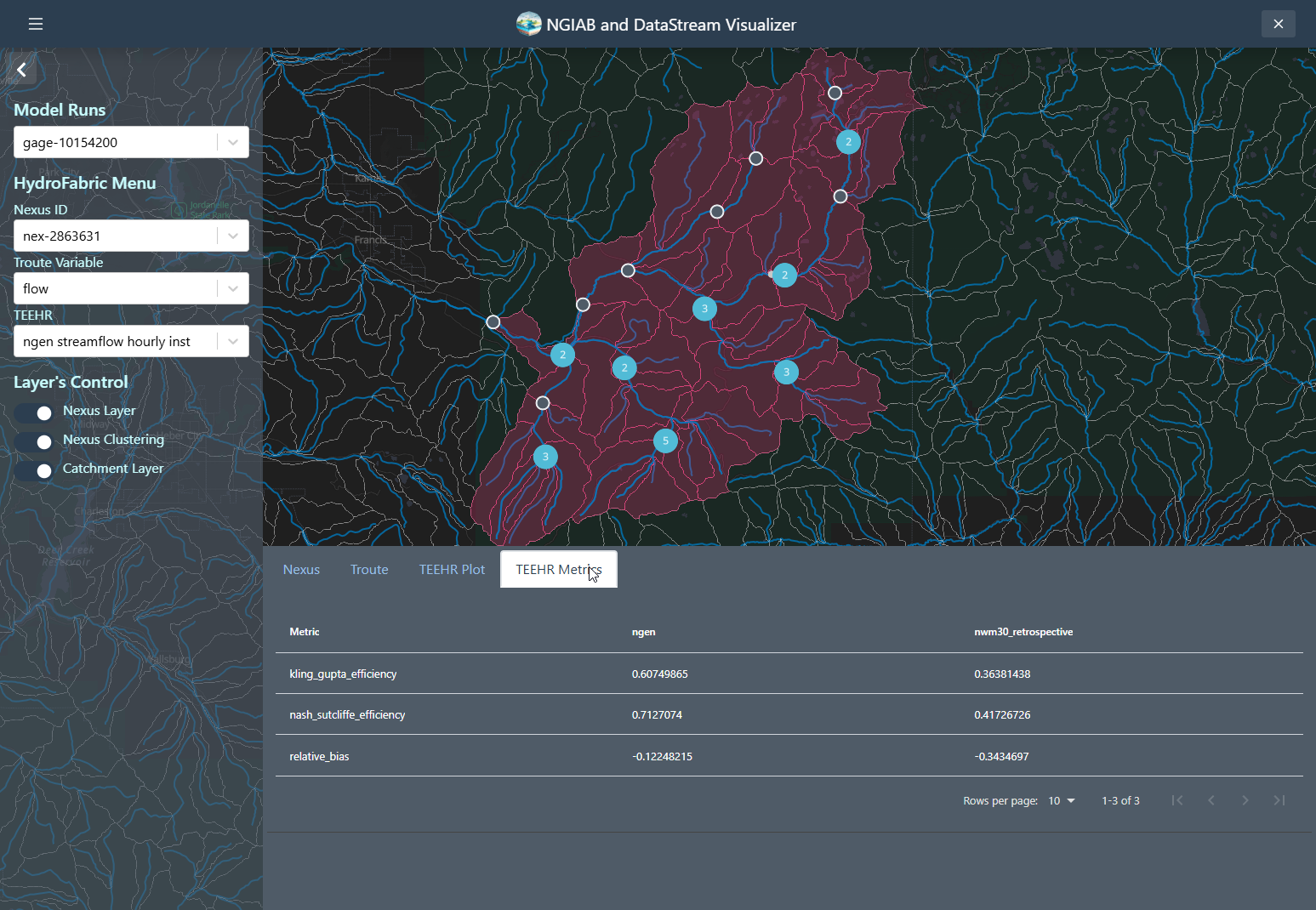
Similarly, a TEEHR evaluation metric can be visualized by going to the metrics tab
Using Data Visualizer with SSH
To use the Data Visualizer through an Secure Shell (SSH) connection, you will have to set up port forwarding when connecting to the remote machine. Port forwarding will allow you to access a remotely hosted browser session on your local machine. See the instructions under “Using NGIAB through an SSH connection” in the Advanced Topics episode in this training module.
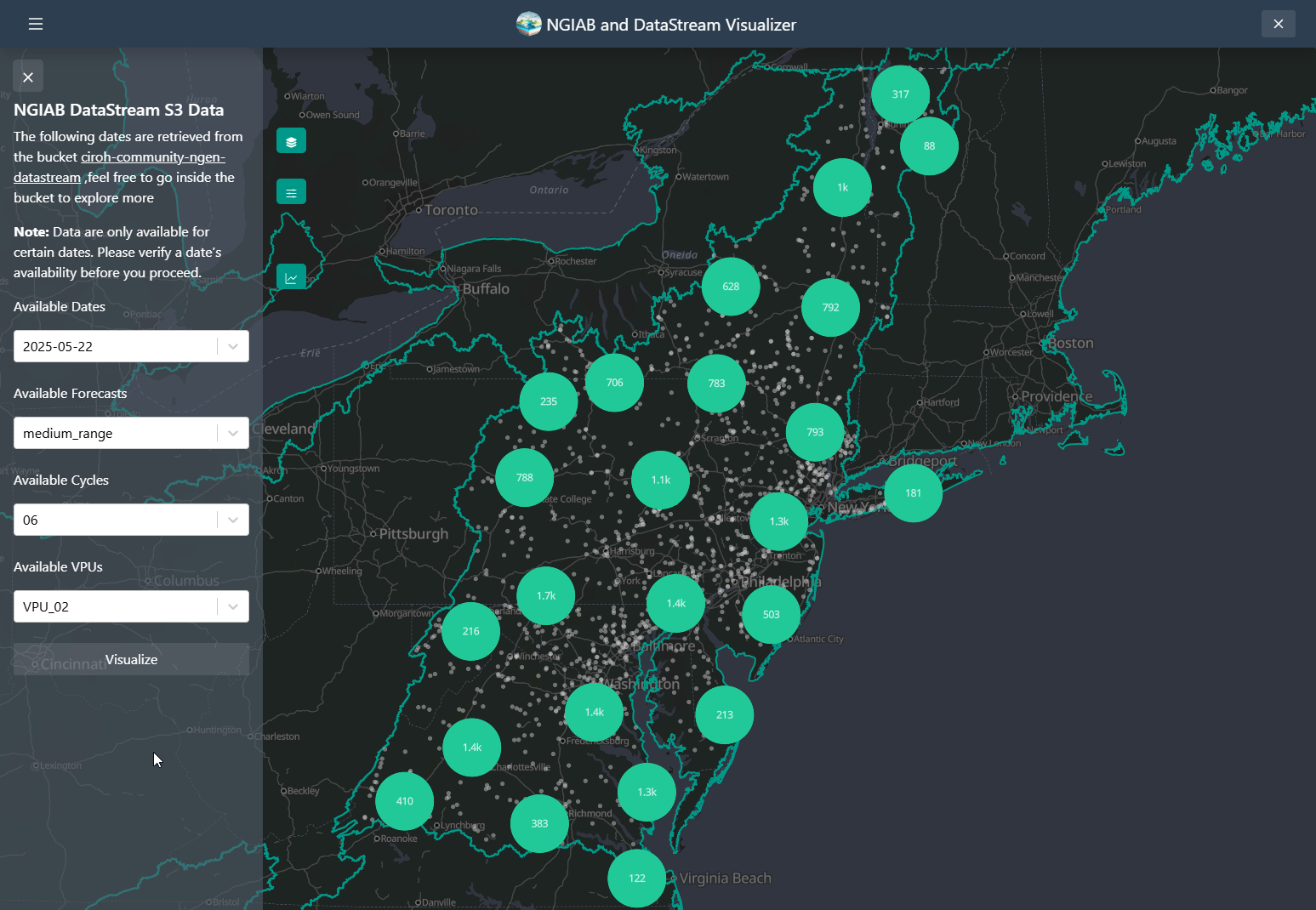
NGIAB DataStream UI
The Visualizer also allows the user to download data as well from an
S3 bucket
containing the output of the NextGen
DataStream. The ViewOnTethys.sh script will create a
~/.datastream_ngiab directory in which it saves all the
different outputs downloaded by the visualizer. It will also create a
~/.datastream_ngiab/datastream_ngiab.json in which metadata
will be saved to locate the downloaded output directories. It serves as
a cache, so it allows the user to look first at the
~/.datastream_ngiab before trying to download the data
BASH
ℹ Reclaiming ownership of /home/aquagio/.datastream_ngiab (sudo may prompt)…
ℹ No existing Datastream cache found – a fresh download will be used.The .datastream_ngiab.json appends the different
downloads with metadata that allows the user to know the file being
downloaded. The prefix belongs to the path on the s3
bucket. The label is created with the following format:
ngen.<date>_<forecast_type>_<cycle>_<VPU>
JSON
{
"datastream": [
{
"label": "ngen.20250522_medium_range_06_VPU_02",
"bucket": "ciroh-community-ngen-datastream",
"prefix": "v2.2/ngen.20250522/medium_range/06/VPU_02/ngen-run.tar.gz",
"path": "/var/lib/tethys_persist/.datastream_ngiab/ngen.20250522_medium_range_06_VPU_02",
"date": "2021-01-01:00:00:00",
"id": "15145d327f19426b890e4465160f963a"
}
]
}Note assuming only the first ensemble. If we are
specific it will look like this:
ngen.<date>_<forecast_type>_<cycle>_<ensemble>_<VPU>
This functionality allows the user to be able to quicklu search the data they want from the S3 bucket containing the output of the NextGen DataStream. They can explore and download as needed.
Your Turn
Use the ViewOnTethys.sh script to launch the Data
Visualizer in Docker, then open
http://localhost:${NGINX_PORT} in your browser. Explore at
least three different visualization modes—such as Nexus time series,
catchment hydrographs, and TEEHR performance metrics. Next, edit
~/ngiab_visualizer/ngiab_visualizer.json to add a new model
run (define its label and path), restart the visualizer, and confirm it
appears in the dropdown menu.
- The Data Visualizer (built on the Tethys Platform) provides interactive geospatial maps and time series plots for NextGen model outputs in NGIAB.
- It integrates seamlessly with NGIAB via
guide.shorViewOnTethys.sh. - Model outputs reside under
~/ngiab_visualizer, with metadata stored inngiab_visualizer.json. - You can visualize Nexus points, catchment summaries, Troute variables, and TEEHR hydrographs and performance metrics.
Content from Calibration
Last updated on 2025-06-10 | Edit this page
Overview
Questions
- How do I calibrate parameters for a NextGen run?
Objectives
- Create calibration configurations for a NextGen run
- Run the
ngiab-calpackage
Parameter Calibration using ngiab-cal
ngiab-cal is a Python package that creates calibration
configurations and copies calibrated parameters to a NextGen model
configuration. It works with the NGIAB folder structure to ensure
compatibility with the other suite of NGIAB tools.
The modeled time period is split into three periods: warmup, calibration, and validation. After the warmup period, the model parameters are adjusted to match observations in the calibration period, and the validation period is used to test the calibrated parameters.
Using ngiab-cal
To use ngiab-cal without installation:
To install ngiab-cal as a package:
A configuration file at calibration/ngen_cal_conf.yaml
controls the calibration process. Users can edit the configuration file
to meet their needs and preferences, such as number of iterations,
acceptable parameter ranges, and time periods. The layout of the
configuration file is as follows:
general:
strategy:
type: estimation
algorithm: dds # Uses Dynamically Dimensioned Search algorithm
name: calib # Don't modify this
log: true # Enable logging
workdir: /ngen/ngen/data/calibration # Don't modify this working directory in the Docker container
yaml_file: /ngen/ngen/data/calibration/ngen_cal_conf.yaml # Don't modify this either
iterations: 100 # Number of calibration iterations (customizable with -i flag)
restart: 0 # Start from beginning (0) or resume from iteration
# Model configurations
CFE:
- name: b # CFE parameter name
min: 2.0 # Minimum allowed value
max: 15.0 # Maximum allowed value
init: 4.05 # Initial value
- name: satpsi
min: 0.03
max: 0.955
init: 0.355
# Additional parameters...
eval_params:
objective: kge # Kling-Gupta Efficiency as objective function
evaluation_start: "..." # Start time for calibration period
evaluation_stop: "..." # End time for calibration period
valid_start_time: "..." # Start time including warmup
valid_end_time: "..." # End time of simulation
# Additional time parameters...
basinID: 01646500 # USGS gage ID
site_name: "USGS 01646500: " # Label for plotsCalibrating and saving parameters requires the three following
commands, where /path/to/ngiab/data/folder is replaced with
the appropriate filepath, and USGS_GAGE_ID is replaced with
an appropriate USGS gage within your study area.
BASH
# Create calibration configuration
ngiab-cal /path/to/ngiab/data/folder -g USGS_GAGE_ID
# Create and run calibration (200 iterations)
ngiab-cal /path/to/ngiab/data/folder -g USGS_GAGE_ID --run -i 200
# Force recreation of calibration configuration
ngiab-cal /path/to/ngiab/data/folder -g USGS_GAGE_ID -fMore details about usage of ngiab-cal can be found on its GitHub page.
Your Turn
Use the ngiab-cal package to calibrate parameters for
the input data that you used for your latest run.
Extra Credit: execute a NextGen run again, and compare the performance between calibrated parameters and uncalibrated parameters.
- The
ngiab-calpackage is used to calibrate parameters for a NextGen model run. -
ngiab-calis a command-line tool controlled via a YAML configuration file that determines parameter ranges, time periods, and evaluation metrics.
Content from Advanced Topics
Last updated on 2025-05-01 | Edit this page
Overview
Questions
- How do I use NGIAB on an high-performance computing (HPC) system?
- How do I use the Data Visualizer through an SSH connection?
- How can I contribute to NGIAB?
Objectives
- Install and use NGIAB on an HPC
- Use port forwarding to view NGIAB results
- Explain the NGIAB community contribution process
The most up-to-date information on installing NGIAB on an HPC can be
found on CIROH’S
NGIAB HPC GitHub page. Other than a different installation process
and the use of Singularity instead of Docker, the workflow is the same
to execute a NextGen run in NGIAB. Tools like the Data Preprocessor,
TEEHR, and the Data Visualizer are still available. The NGIAB-HPCInfra
contains its own interactive guide.sh script, which allows
users to specify input data pathways and run configurations (serial or
parallel), as well as trigger the execution of TEEHR and the Data
Visualizer.
Singularity
NGIAB uses Singularity as its containerization platform for HPC environments. Singularity enables secure execution of containerized applications on multi-user HPC clusters. Key features of Singularity include:
- Native HPC integration, which allows the execution of containerized applications within existing batch job schedulers such as SLURM (Simple Linux Utility for Resource Management) workload manager, PBS (Portable Batch System) and LSF (Load Sharing Facility)
- Enforced security – it runs containers as non-root users, reducing security risks; and
- Access to host file systems – it enables users to interact with datasets and computational resources without additional configuration directly.
This section explains how to run NextGen In A Box (NGIAB) using Singularity on the Pantarhei HPC system at the University of Alabama. To access Pantarhei, please follow the instructions on CIROH’s DocuHub page.
1. Log Into Pantarhei
Open a terminal and connect to the login node:
Replace <USERNAME> with your actual Pantarhei
username.
2. Request a Compute Node (Do NOT run on login node)
On the login node, request an interactive session:
Use the normal partition unless you require more time or
special resources.
5. Download Sample Dataset
Pick one of the sample datasets to download and extract:
Option 1: AWI-009 (Provo River, UT)
BASH
wget https://ciroh-ua-ngen-data.s3.us-east-2.amazonaws.com/AWI-009/AWI_16_10154200_009.tar.gz
tar -xf AWI_16_10154200_009.tar.gzOther options: AWI-007 or AWI-008 can be used similarly, see the Installation and Setup episode.
6. Clone the NGIAB-HPCInfra Repository
✅ Note: Always run
guide.shfrom inside theNGIAB-HPCInfrafolder.
7. Run guide.sh
Make the script executable if needed:
Then run it:
Follow the prompts:
When asked “Do you want to use the same path?”, type
nThen enter the full absolute path to your extracted dataset folder. Example:
This folder must contain:
forcings/
config/
outputs/
The script will:
Detect system architecture
Pull the correct Singularity image
Mount your dataset
-
Allow running in:
Serial mode
Parallel mode
Interactive container shell
NGIAB’s core functions work through an SSH connection without port forwarding. However, to use the Data Visualizer, you will have to set up port forwarding to view visualization results on your local machine’s browser.
To do so, run the following command on your local machine:
Replace username@remote_host with your credentials.
Now, you should be able to run NGIAB as usual through your SSH tunnel, and access Data Visualizer results in your local browser.
The most up-to-date guidelines on community contributions for each repository can be found on its respective GitHub page.
General contribution guidance
- You can use the issue tracker on GitHub to suggest feature requests, report bugs, or ask questions.
- You can change the codebase through Git:
- Create a fork
- Clone the repository locally
- Keep the fork and clone up-to-date
- Create branches when you want to contribute
- Make changes to the code
- Commit to your local branch
- Push commits to your GitHub fork
- Create a pull request when the changes are ready to be incorporated
Your Turn
Based on your own interests and use cases, try out some of these options:
- Install and use NGIAB on your HPC environment
- Use NGIAB through an SSH connection
- Contribute to NGIAB/NextGen!
- NGIAB supports HPC environments through Singularity, not Docker, but the workflow mirrors the local Docker use.
- Port forwarding is required to use the Data Visualizer through an SSH connection.
- Community contribution guidelines are available in each repository’s GitHub page.
